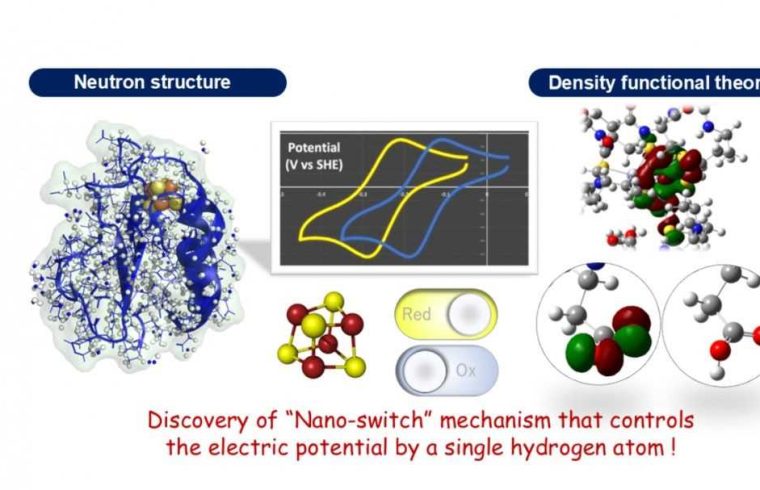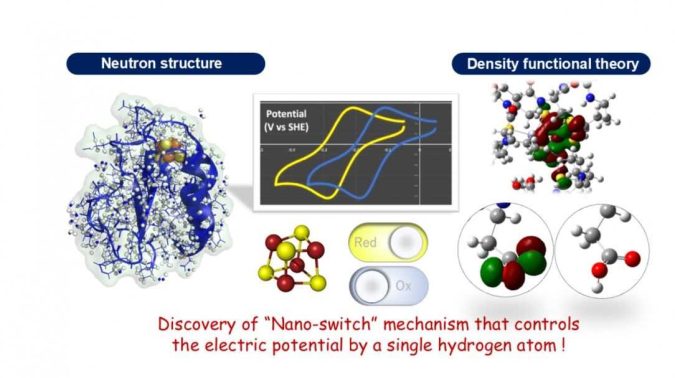
A group of Japanese researchers has discovered, for the first time, a mechanism for adjusting the potential of an “electron carrier” protein in the redox reaction that all organisms use to obtain energy.
Based on experiments, the precise 3D structure of the protein, including hydrogen atoms, was determined, and theoretical calculations using this data visualized the electronic structure of the iron-sulfur cluster. The results revealed, for the first time, that the electric potential of the iron-sulfur cluster changes dramatically depending on the presence or absence of a single hydrogen atom at an amino acid side chain, a so-called “nano-switch” mechanism.
The results will not only deepen our scientific understanding of biological reactions but also provide a major clue to the future development of ultra-sensitive sensors for oxygen, nitric oxide, and novel drugs.
Most reactions in living organisms involve the “electrons” transfer, which is called redox reaction. For example, respiration and photosynthesis can be classified as redox reactions. Some proteins that assist in the electron transfer contain iron and sulfur.
Ferredoxin is a small protein that holds iron-sulfur clusters inside it and is known as the “electron carrier” in living organisms. Ferredoxin is a universal protein that is thought to be present in almost all living organisms; however, the mechanism by which ferredoxin stably carries electrons has remained a mystery to date.
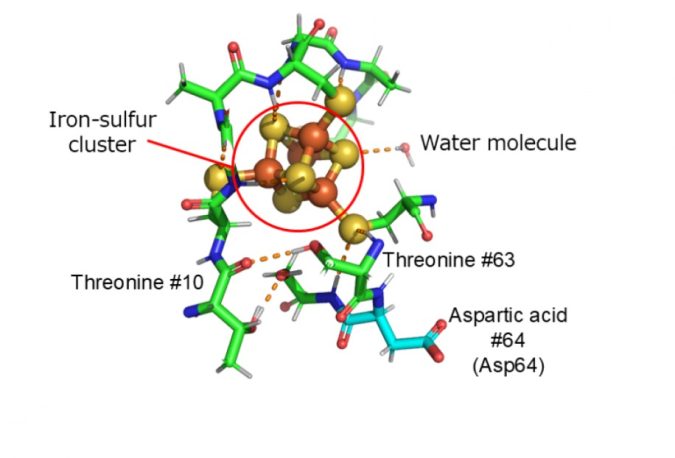
In this study, we conducted experiments using the Ibaraki Biological Crystal Diffractometer (iBIX) at the Materials and Life Science Experimental Facility (MLF) in the Japan Proton Accelerator Research Complex (J-PARC), and have succeeded in determining the precise three-dimensional structure of a ferredoxin at the hydrogen atomic level in experiments using a neutron beam. Visualizing hydrogen atoms in protein molecules using neutrons is extremely difficult, and only less than 0.2% of the entire protein three-dimensional structure database (Protein Data Bank; PDB) has been reported.
Theoretical calculations using experimental geometry including hydrogen atoms were performed to elucidate the electronic structure of the iron-sulfur cluster in the ferredoxin. As a result, it was revealed, for the first time, that an amino acid residue (aspartic acid 64) located far from the iron-sulfur cluster has a significant effect on probability of electron transfer in the iron-sulfur cluster, and plays a role like a switch that controls the electron transfer in ferredoxin. Furthermore, it was shown that the mechanism is universal in various organisms.
The results will not only deepen our scientific understanding of biological reactions but also provide a major clue to the future development of ultra-sensitive sensors for oxygen, nitric oxide, and novel drugs.
This study was published in the online edition of the international scientific journal eLife on November 15, 2024 (Reviewed Preprint).
Lead researchers: Professor Yasutaka Kitagawa of Osaka University, Professor Kei Wada of Miyazaki University, and Professor Masaki Unno of Ibaraki University in collaboration with researchers from Tokyo University of Pharmacy and Life Sciences, Kurume University, Comprehensive Research Organization for Science and Society (CROSS), and Japan Synchrotron Radiation Research Institute (JASRI).