Flooding is a global risk, according to the World Bank, with the lives and property of billions of people threatened. Even more, people are at risk of starvation as a knock-on effect of floods: the waters can drown crops.
Now, researchers are getting closer to identifying the molecular processes underlying how floods deprive plants of oxygen — and how to engineer hardier crops.
Through a meta-analysis, which involves re-analyzing data from other studies en masse, the team from Hiroshima University’s Graduate School of Integrated Sciences for Life uncovered several common genes and their related mechanisms in rice (Oryza sativa) and thale cress (Arabidopsis thaliana).
They published their results on July 19 in Life.
“Hypoxia is an abiotic stress for plants often caused by flooding,” said first author Keita Tamura, referring to the oxygen deprivation caused by oversaturation. “Although many studies have been performed previously, we thought hidden biological mechanisms might be found by analyzing multiple studies through a meta-analysis of publicly available data.”
The team focused on rice and thale cress since the genetics of both have been extensively studied, providing ample amounts of data. Rice is also considered one of the world’s most important crops, serving as a staple food for more than four billion people, according to the Consultative Group for International Agricultural Research, so understanding how to prevent a hypoxia response in the plant is critical, Tamura said.
The researchers identified 29 pairs of RNA-sequencing data for thale cress and 26 pairs for rice for the plants in both normal oxygen and oxygen-deprived states from the available datasets. RNA-sequencing involves transcribing the genetic blueprints of the subject in a specific moment, meaning the data can be used to investigate which genes triggered which changes, according to corresponding author Professor Hidemasa Bono.
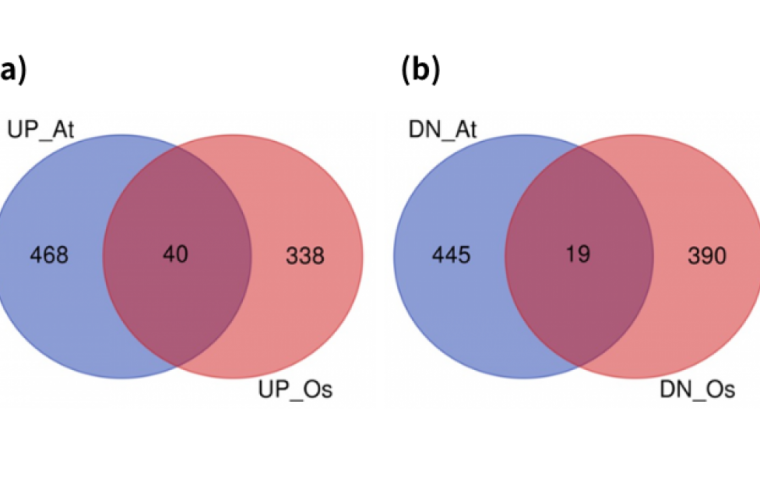
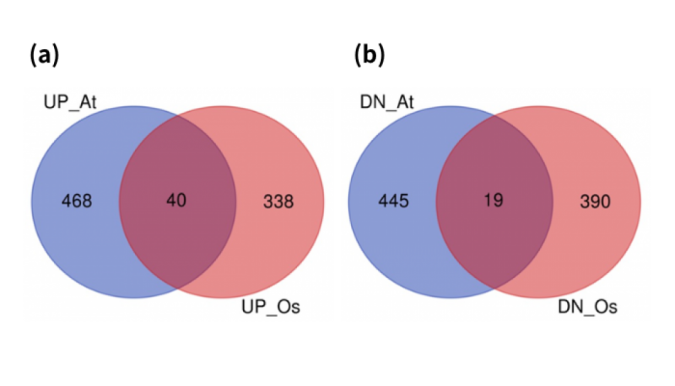
“By analyzing RNA-sequencing data of hypoxia treatments in thale cress and rice, we identified 40 and 19 commonly upregulated and downregulated genes in both species,” Bono said. “Among them, some WRKY transcription factors and cinnamate-4-hydroxylase, whose role in hypoxia response remain unknown, were commonly upregulated in both thale cress and rice.”
According to Bono, this common upregulation means that these molecular machineries became more active under oxygen deprivation, indicating they have specific mechanistic responsibilities for how the plants respond.
Bono and Tamura compared their results to a similar meta-analysis of hypoxia in human cells and tissue samples. They found two of the commonly upregulated genes in rice and thale cress were downregulated in their human counterparts.
“Our meta-analysis suggests distinct molecular mechanisms under hypoxia in plants and animals,” Bono said. “The candidate genes identified in this study are expected to elucidate novel molecular mechanisms of hypoxia responses in plants. Ultimately, we plan to manipulate one of the candidate genes through genome editing technology to create flood-tolerant plants.”