Researchers at Kanazawa University report in Nature Communications the discovery that in the developing fly brain, neurons stemming from the same parent cell experience repulsion. This lineage-dependent repulsion is regulated by a protein known as Dscam1.
The brain’s structure has columnar features, which are hypothesized to arise from nerve cells (neurons) stemming from the same parent cell, initially forming radial units. How exactly this process unfolds at the molecular level remains unexplained, however.
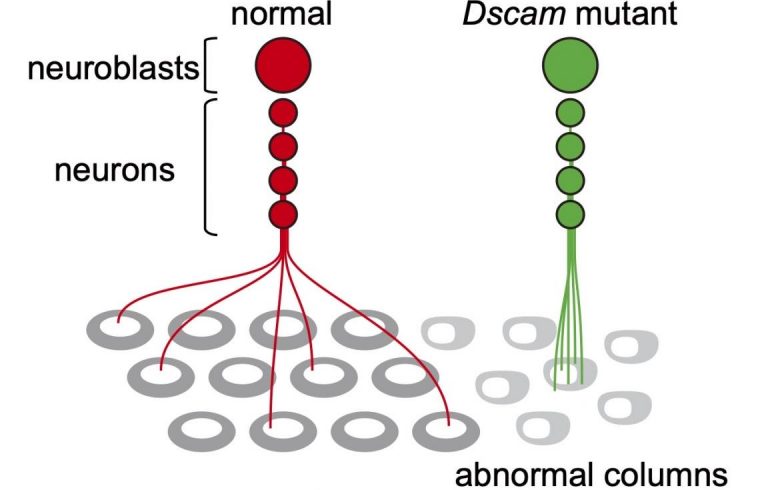
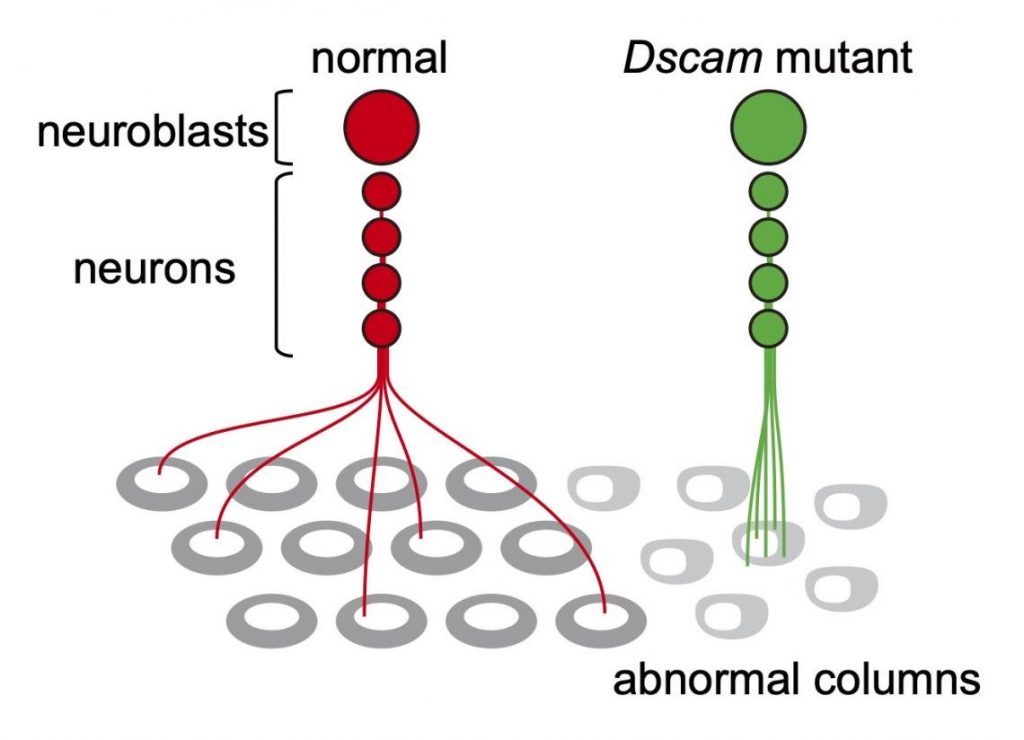
Now, an important insight comes from Makoto Sato and colleagues from Kanazawa University who show how, in the fly brain, a gene known as Dscam regulates how neurons from one lineage repel each other, and project their axons to different columns. (Axons or nerve fibers are long protrusions of nerve cells, the function of which is to conduct electrical signals.) This finding corroborates the ‘radial unit hypothesis’, with the mechanism at play being lineage-dependent repulsion between sister neurons.
The researchers first looked at the evolution of neuron growth in the medulla, a part of the fly’s visual system featuring a columnar structure. Its development is similar to that of the cerebral cortex in the brain of mammals; it involves neuroblasts (neural stem-like cells) that produce radially oriented and clonally related groups of neurons.
Sato and colleagues recorded the distances between sister neurons (i.e., neurons stemming from the same neuroblast and forming a radial unit) and between axon pairs. From the obtained distance data, the scientists were able to conclude that the sister neurons often repel each other — this observation is consistent with the formation of columns. Sato and colleagues call this process ‘lineage-dependent repulsion’.
The mechanism that enables lineage-dependent repulsion must lie in daughter neurons derived from the same neuroblast ‘remembering’ the identity of their common mother neuroblast. Sato and colleagues put forward the explanation that the protein Dscam1 is involved.
Dscam1 can develop nearly 20,000 variants, but when two identical Dscam1 molecules bind, they lead to a repulsive signal known to control self-avoidance in certain dendritic processes — dendrites are branch-like extensions of nerve cells. The reasoning then is that daughter neurons stemming from the same neuroblast produce the same Dscam1 variant, and so repel each other, whereas neurons of different lineages express different Dscam1 variants that don’t repel each other and can project to the same column.
The scientists were able to support their argumentation by a series of experiments confirming the relation between Dscam1 and lineage-dependent repulsion. Sato and colleagues note that “the mechanism that we propose … is very simple”, and add that it will be “interesting to determine whether similar mechanisms exist in other biological systems including column formation in mammalian brains.”